This is the latest version of "SALAD on ARRAYs". (Last update 2011-05-18)
For full functionality and display we recommend you use FireFox version 3.0 or later
We welcome your questions and comments.
Old version (pollen, rice) is available here.
For full functionality and display we recommend you use FireFox version 3.0 or later
We welcome your questions and comments.
Old version (pollen, rice) is available here.
|
1)Search by annotated protein information 2)Search by Blast search results for your sequences 3)Search by Pfam domain information 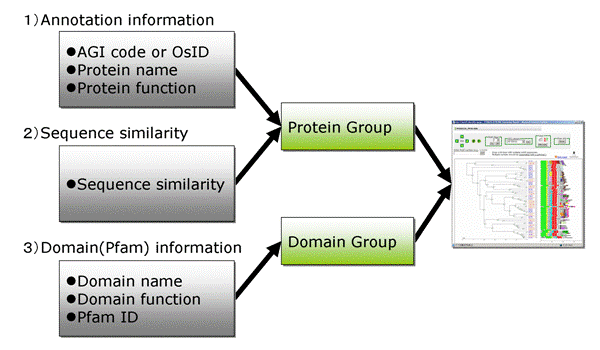
| |
The following systems are needed in order to browse this page.
| Windows XP sp2 Windows VISTA |
・Firefox 3.0 or later(recommend) ・Internet Explorer 6.0 or later(exclude IE8) (If you use Internet Explorer 8, please turn on the "Compatibility View" to fix the problem. ) |
| Macintosh OS X 10.2 or later |
・Firefox 3.0 or later(recommend) ・Safari 4.0 or later |
About Group
SALAD clustering data was created for each annotation group. When
clustering protein sequences in an annotation group, certain SALAD
motifs were extracted as conserved amino acid sequence patterns, those
occur repeatedly in the group. Therefore, the extracted motif set may be
distinct from that extracted from other related groups. To avoid
misunderstanding clustering data and get more new insights, we recommend
you to analyze several groups with the same annotation of your interest.
Since SALAD database also has a function for interactive SALAD analysis,
you can try SALAD clustering analysis for your own annotation group by
uploading your multifasta file.
 SALAD on ARRAYs HELP
SALAD on ARRAYs HELP