Blasting SALAD Analysis HELP (English)
Table of contents
- Summary
- Data
- How to use
New Function Addition (updated on 2012.05.14)
You can download sequence file(fasta format).
|
< procedure >
Click any node numbers in a SALAD data window, you can get all the sequence information which are contained in the selected node.
*Couldn't select each annotation to get each sequence information
|
1. Summary
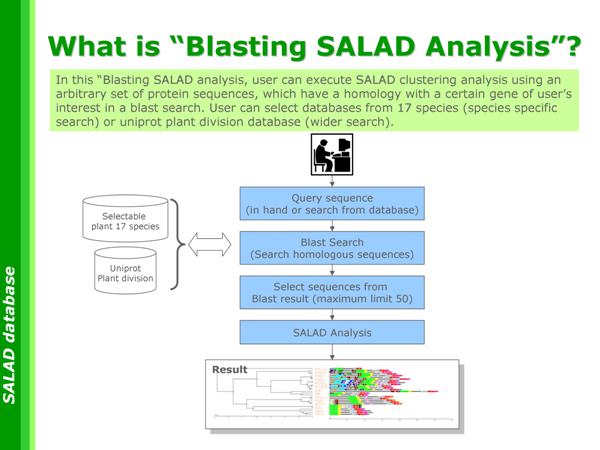
What is "Blasting SALAD Analysis"?
Basic precautions
- Restriction
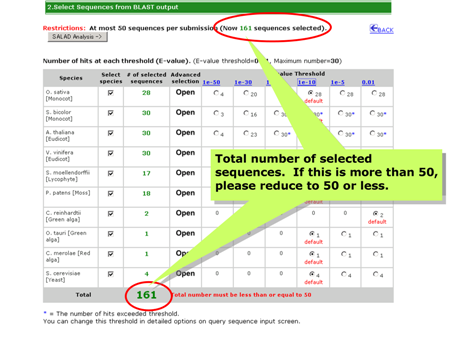
Because of the performance of our machine, the number of sequences in a dataset is limited to at most 50 per submission. User may need to select sequences from the blast result by yourself because each dataset is set according to the purpose of analysis. When you select sequence, please be careful not to exclude any important sequences. We provide the page below, which contains helpful information for this selection. Please take full advantages of it.
2. Data
Blast Database
There are two databases available:
- Selectable 17 species [for species specific search]
- Uniprot (Swiss-Prot/TrEMBL) plant division [for wider search]
In uniprot (TrEMBL), isoforms and variants encoded by the same gene are
stored in separate entries. For example, there are three COI1 sequences
of Arabidopsis thaliana (O04197, B2BDA3 and B2BD96) in uniprot and the
similarity is more than 99% among them. To remove this redundancy, we
marge sequences using cd-hit on the basis of
identity (>= 98%) and coverage (>= 95%).
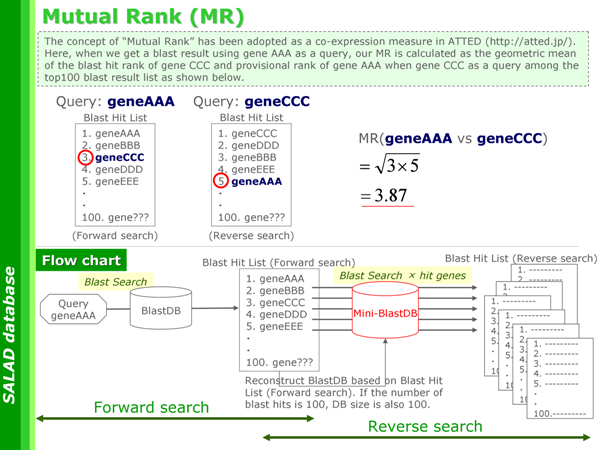
MR
|
MR | ... | This value is calculated as geometric mean of the blast hit rank of B when A as a query and that of A when B as a query.
|
|
Intraspecific-MR | ... | This value is calculated as geometric mean of the blast hit within-species rank of B when A as a query and that of A when B as a query.
|
3. How to use
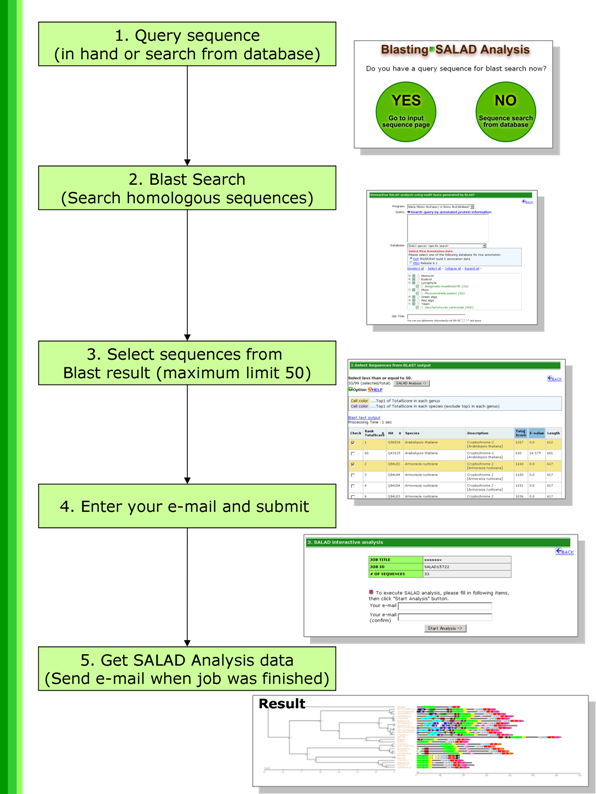
Procedure
How to select sequences from blast result
Depending on the databases for blast search, the selection pages are different.
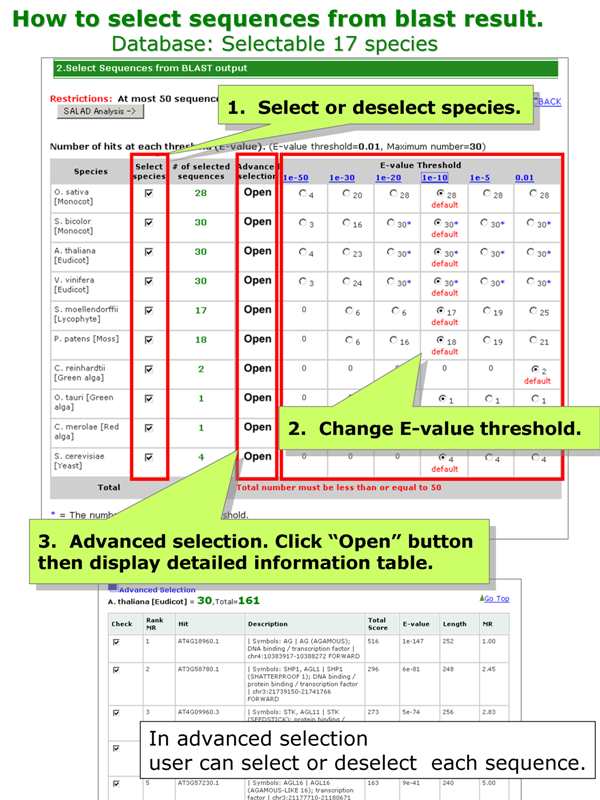
[Selectable 17 species]
Check the total number of selected sequences!
There are three methods to select or deselect sequence.
- Select or deselect species
- Change E-value threshold
- Advanced selection
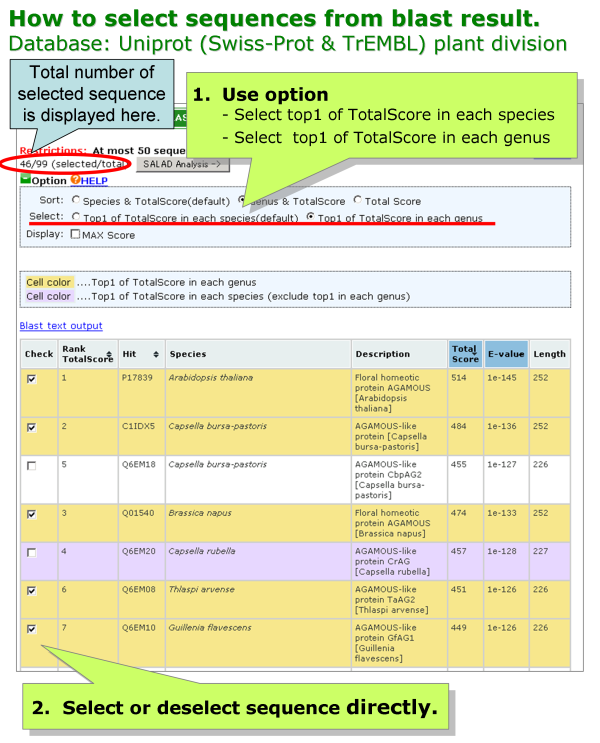
[Uniprot (Swiss-Prot & TrEMBL) plant division]
There are two methods to select or deselect sequence.
- Use option
- Select or deselect sequence directly
Detailed options for blast search
Criteria for calculating MR
| Total score | ... |
By the sum of scores from all HSPs from the same database sequence |
| Max score | ... |
By the bit score of HSPs, similar to Expect Value |
| E-value | ... |
Expect Value |
| Query coverage | ... |
By the percent of length coverage for the query |
Caution
Caution
- When you select blastp in "Blast Search" step, "Add query" option will be displayed.("Add query" option isn't displayed when you select blastx in "Blast Search" step.)
- When your query sequence isn't in FASTA format containing header(query name), "User Query" will be added as a query name.
|